Mastering Single-Cell RNA-Seq Analysis in R - Post 10: Why Your Clustering Algorithm Choice Matters! Link to heading
Ever wondered why some researchers get clean cell type clusters while others struggle with messy, overlapping groups? After creating beautiful UMAP visualizations in Post 9.5, you face the critical challenge of defining discrete cell clusters for quantitative analysis.
Today we’re diving into Louvain vs Leiden clustering algorithms - and why making the right choice could revolutionize the quality and reproducibility of your single-cell discoveries!
🎯 The Clustering Challenge: From Continuous to Discrete Link to heading
The Fundamental Problem Link to heading
UMAP creates beautiful continuous visualizations where cells flow smoothly across 2D space, but biological analysis requires discrete categories. You need to draw boundaries that separate your continuous cellular landscape into distinct, analyzable groups.
The Mathematical Challenge:
- Input: Continuous high-dimensional relationships between cells
- Output: Discrete cluster assignments for each cell
- Goal: Boundaries that reflect genuine biological distinctions, not arbitrary mathematical divisions
Why Clustering Algorithm Matters Link to heading
All clustering isn’t created equal. The algorithm you choose determines:
- Boundary quality - How well clusters separate genuine cell types
- Stability - Whether you get consistent results across runs
- Biological interpretability - How easily clusters map to known cell types
- Downstream analysis success - Quality of differential expression and cell type annotation
The same dataset with the same parameters can produce dramatically different results depending on your algorithmic choice.
🥈 Louvain Algorithm - The Historical Standard Link to heading
The Community Detection Pioneer Link to heading
Louvain clustering, developed for social network analysis, became the default choice for single-cell clustering due to its speed and reasonable performance. Most researchers use it without realizing there are superior alternatives.
How Louvain Works:
- Initialization: Each cell starts as its own cluster
- Local optimization: Iteratively move cells to neighboring clusters if it improves modularity
- Aggregation: Merge clusters and repeat until no improvement
- Termination: Stop when no beneficial moves exist
The Fundamental Flaw: Local Optima Traps Link to heading
The Problem:
Louvain can get trapped in “locally optimal” solutions that aren’t globally optimal. Once it finds a clustering that’s better than small modifications, it stops searching - even if dramatically better solutions exist.
Real-World Impact:
- Suboptimal boundaries between cell types
- Artificially split populations that should be unified
- Merged populations that should be separated
- Inconsistent results across different runs or parameter values
Performance Characteristics Link to heading
Strengths:
- Fast execution on large datasets
- Wide adoption and community familiarity
- Good enough results for many applications
- Stable implementation in most software packages
Limitations:
- Gets trapped in suboptimal solutions
- Less stable results across random seeds
- Poorer quality cluster boundaries
- Reduced reproducibility in complex datasets
🏆 Leiden Algorithm - The Superior Solution Link to heading
The Algorithmic Innovation Link to heading
The Leiden algorithm, published in 2019, specifically addresses Louvain’s fundamental limitations while maintaining computational efficiency. It represents a genuine algorithmic advancement, not just an incremental improvement.
Links: Leiden Paper | leidenalg Python Package
How Leiden Escapes Local Optima Link to heading
The Innovation:
Leiden adds a crucial “refinement” phase that prevents entrapment in local optima:
- Standard optimization like Louvain
- Partition refinement - Subdivide clusters and re-optimize
- Quality verification - Ensure no further improvements possible
- Iterative improvement until global optimum reached
The Result:
Leiden keeps searching until it finds the truly best clustering, not just one that’s better than minor modifications.
Superior Performance Characteristics Link to heading
Better Optimization:
- Escapes local optima that trap Louvain
- Finds higher-quality cluster boundaries
- Achieves better modularity scores consistently
- More thorough optimization of cluster assignments
Enhanced Stability:
- Reproducible results across multiple runs
- Consistent clustering with different random seeds
- Robust performance across parameter ranges
- Reliable boundaries between cell populations
Biological Advantages:
- Cleaner cell type separation with fewer mixed populations
- More interpretable clusters that map clearly to known biology
- Better differential expression due to purer cell populations
- Improved downstream analysis across all applications
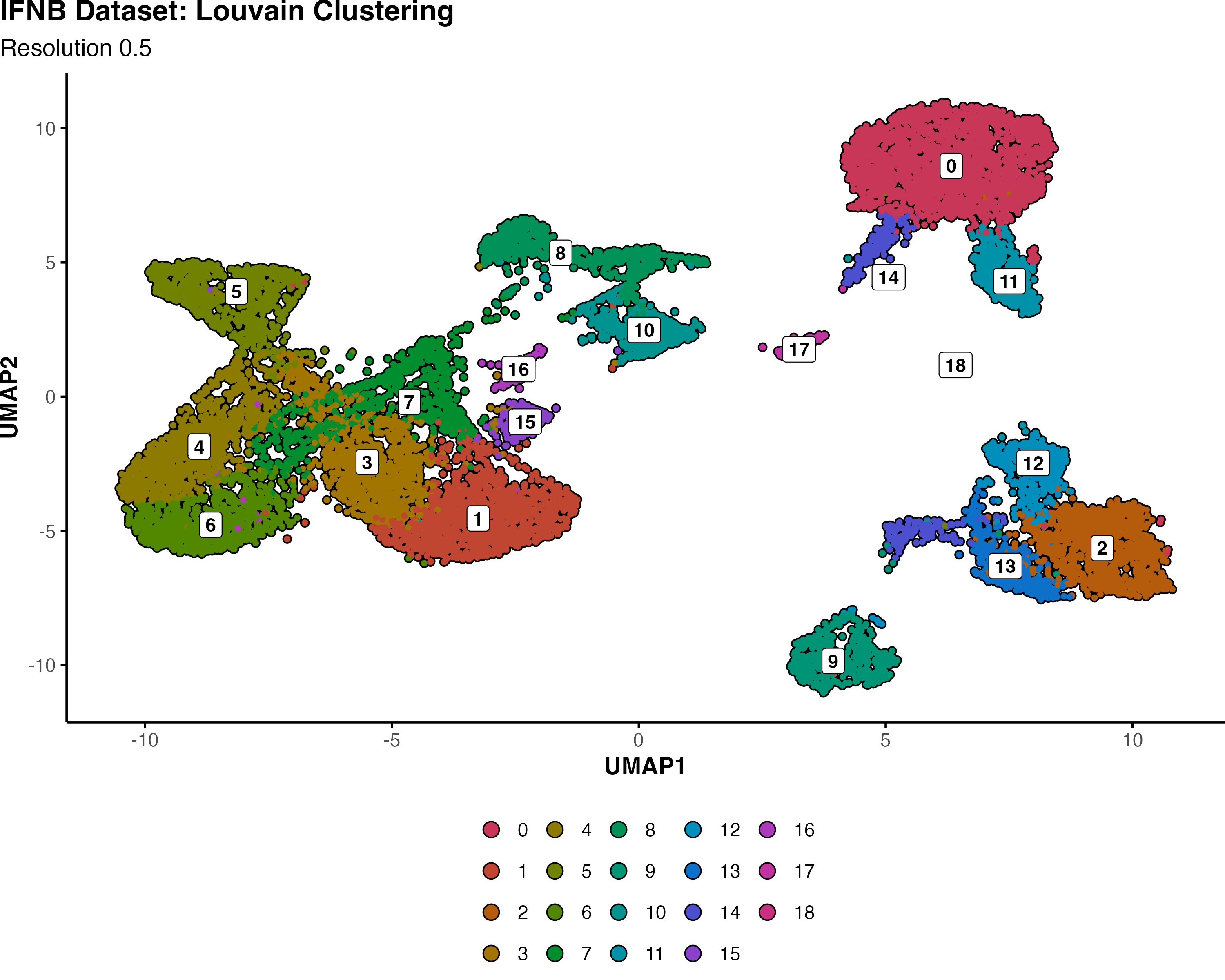
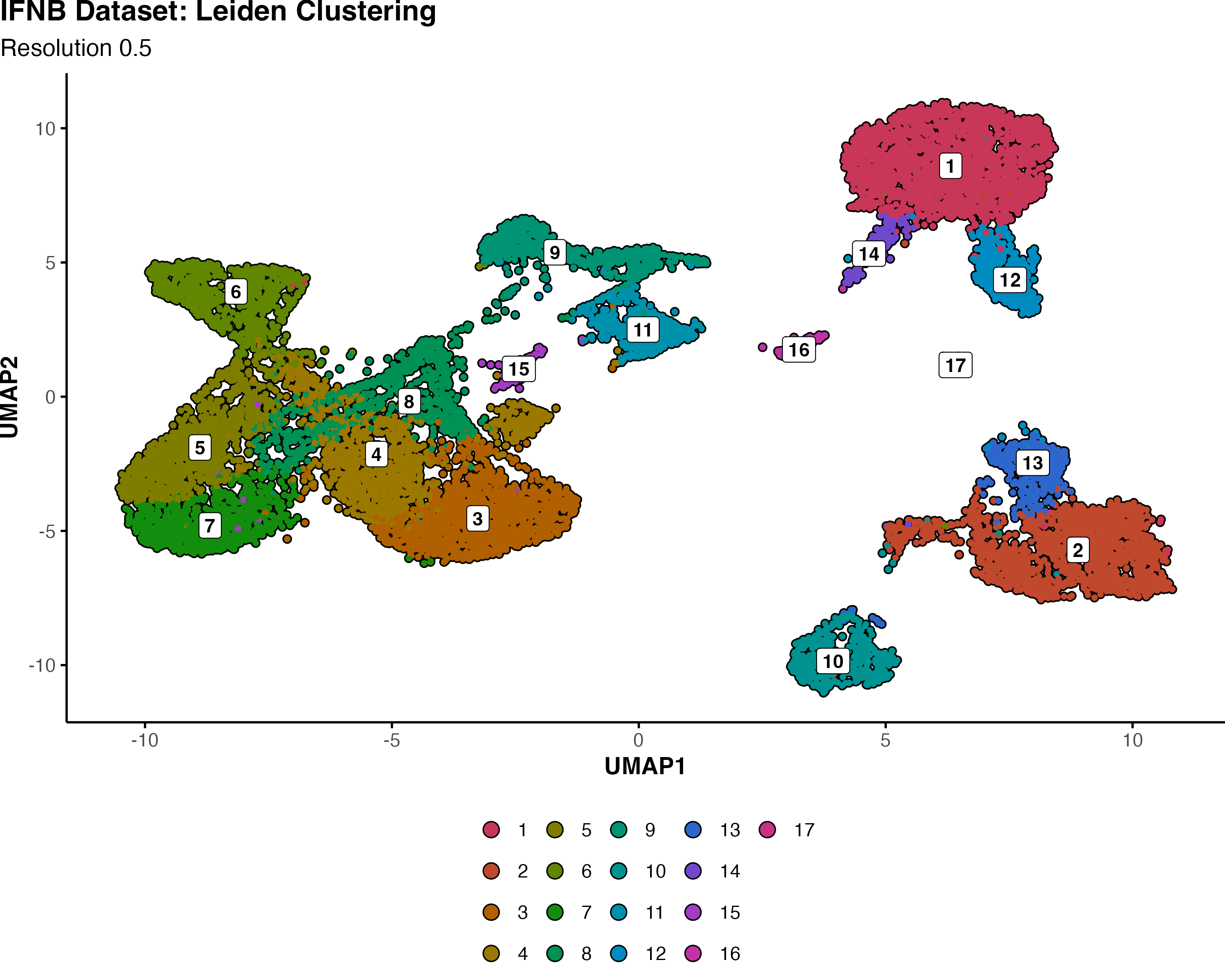
🔬 Real-World Comparison: IFNB Dataset Analysis Link to heading
The Experimental Setup Link to heading
Using the interferon-stimulated PBMC dataset at resolution 0.5, we can directly compare how Louvain and Leiden handle the same biological data:
library(Seurat)
# First, build the neighbor graph (same for both algorithms)
ifnb <- FindNeighbors(ifnb,
reduction = "pca",
dims = 1:30) # Use 30 PCs based on elbow plot
# Louvain clustering (Seurat default)
ifnb_louvain <- FindClusters(ifnb,
resolution = 0.5,
algorithm = 1) # Louvain
# Leiden clustering (recommended)
ifnb_leiden <- FindClusters(ifnb,
resolution = 0.5,
algorithm = 4) # Leiden
The Quantitative Results Link to heading
Louvain Clustering:
- 19 total clusters (labeled 0-18)
- More granular subdivision of cellular populations
- Several small clusters that may represent over-splitting
- Some potential spurious separations
Leiden Clustering:
- 17 total clusters (labeled 1-17)\
- More consolidated, biologically coherent groupings
- Merged some of Louvain’s smaller clusters into unified populations
- Cleaner separation between major cell types
The Biological Interpretation Link to heading
Why Fewer Clusters Can Be Better:
The reduction from 19 to 17 clusters isn’t a loss of resolution - it’s an improvement in biological accuracy. Leiden’s superior optimization identified that some of Louvain’s boundaries were artificial artifacts rather than genuine biological distinctions.
Enhanced Cell Type Identification:
- Cleaner populations make cell type annotation more straightforward
- Reduced mixed clusters decrease annotation ambiguity\
- Better marker gene specificity due to purer populations
- Improved downstream analysis across all applications
💪 Implementation: Making the Switch Link to heading
The Critical Parameter Link to heading
Important: Seurat still defaults to Louvain! You must explicitly request Leiden clustering:
# Recommended: Use Leiden algorithm
FindClusters(ifnb,
resolution = 0.5,
algorithm = 4) # Leiden
# Default (not recommended): Louvain algorithm
FindClusters(ifnb,
resolution = 0.5) # algorithm = 1 (Louvain)
The Complete Workflow Link to heading
# Complete clustering workflow with Leiden
ifnb <- ifnb %>%
# Build neighbor graph from PCA space
FindNeighbors(reduction = "pca", dims = 1:30) %>%
# Apply Leiden clustering
FindClusters(resolution = 0.5, algorithm = 4) %>%
# Generate UMAP coordinates
RunUMAP(dims = 1:30)
# Visualize results
DimPlot(ifnb, reduction = "umap", label = TRUE)
Performance Improvements Link to heading
Historical Context:
Early implementations of Leiden in Seurat were slower than Louvain, creating hesitation about adoption. Recent improvements have eliminated this performance gap.
Current Reality:
- Computational efficiency now comparable to Louvain
- Memory usage similar to traditional methods
- Execution time acceptable for standard datasets
- No practical barriers to routine adoption
🧠 Why the Algorithm Choice Matters for Everything Link to heading
Downstream Analysis Quality Link to heading
Differential Expression:
Purer clusters from Leiden produce cleaner differential expression results:
- Stronger statistical signals due to reduced population mixing
- More specific marker genes for each cluster
- Better biological interpretability of expression differences
- Higher validation rates in follow-up experiments
Cell Type Annotation:
Leiden’s cleaner boundaries make annotation more straightforward:
- Less ambiguous cluster identities
- Clearer correspondence to known cell types
- Reduced manual curation needed
- More confident biological conclusions
Trajectory Analysis:
Better clustering improves developmental analysis:
- Cleaner starting points for trajectory inference
- More accurate transition detection between states
- Better biological interpretation of developmental processes
Reproducibility Benefits Link to heading
Consistent Results:
Leiden’s stability advantages compound across your analysis pipeline:
- Same clustering across multiple runs
- Reliable results for collaborators
- Reproducible publications that others can validate
- Stable foundations for method development
Cross-Laboratory Validation:
When other groups analyze similar data:
- Consistent cell type identification across studies
- Comparable biological conclusions between laboratories\
- Meta-analysis compatibility across datasets
- Scientific credibility through reproducible findings
🔥 The Strategic Impact Link to heading
The Quality Cascade Link to heading
Your clustering algorithm choice creates a quality cascade that affects every subsequent analysis:
Good Clustering (Leiden) → Pure Populations → Strong DE Signals → Clear Biology → Reproducible Discoveries
Poor Clustering (Louvain) → Mixed Populations → Weak DE Signals → Confusing Biology → Irreproducible Results
The Time Investment Link to heading
Short-term: Zero additional effort - just change one parameter
Long-term: Months saved from cleaner analysis and fewer artifacts
The switch from Louvain to Leiden requires no additional learning, no new software, and no workflow changes. It’s purely an algorithmic upgrade with immediate benefits.
The Professional Standard Link to heading
As the single-cell community recognizes Leiden’s advantages, using Louvain increasingly signals:
- Outdated methodology
- Lack of awareness of algorithmic improvements
- Suboptimal analysis quality
- Reduced professional credibility
Adopting Leiden demonstrates:
- Current best practices
- Attention to methodological advances
- Commitment to analysis quality
- Professional competence
🎉 The Bottom Line Link to heading
Leiden clustering doesn’t just find clusters - it finds better clusters. The algorithmic improvements translate directly into cleaner cell type identification, more reproducible results, and superior biological interpretability.
The choice between Louvain and Leiden represents one of the easiest upgrades you can make to improve your single-cell analysis quality. No new learning required, no additional computational burden, just demonstrably better results through superior algorithms.
When you have access to methods that escape local optima, provide more stable results, and produce cleaner biological boundaries, continuing to use inferior algorithms becomes a choice to accept suboptimal science.
The single-cell field moves quickly, and algorithmic improvements like Leiden represent genuine advances that every researcher should adopt. Your future self - struggling with unclear cluster boundaries or irreproducible results - will thank you for making the switch.
Ready to let superior algorithms discover your cell types with unprecedented clarity and reproducibility?
Next up in Post 11: Resolution Optimization - Finding the perfect cluster granularity for your specific biological questions! 📈